Viral RNA families

Rfam is collaborating with the European Virus Bioinformatics Centre (EVBC), with an aim to provide a comprehensive database for virologists interested in RNA secondary structures.
Learn more about EVBC →
Coronaviridae RNA families
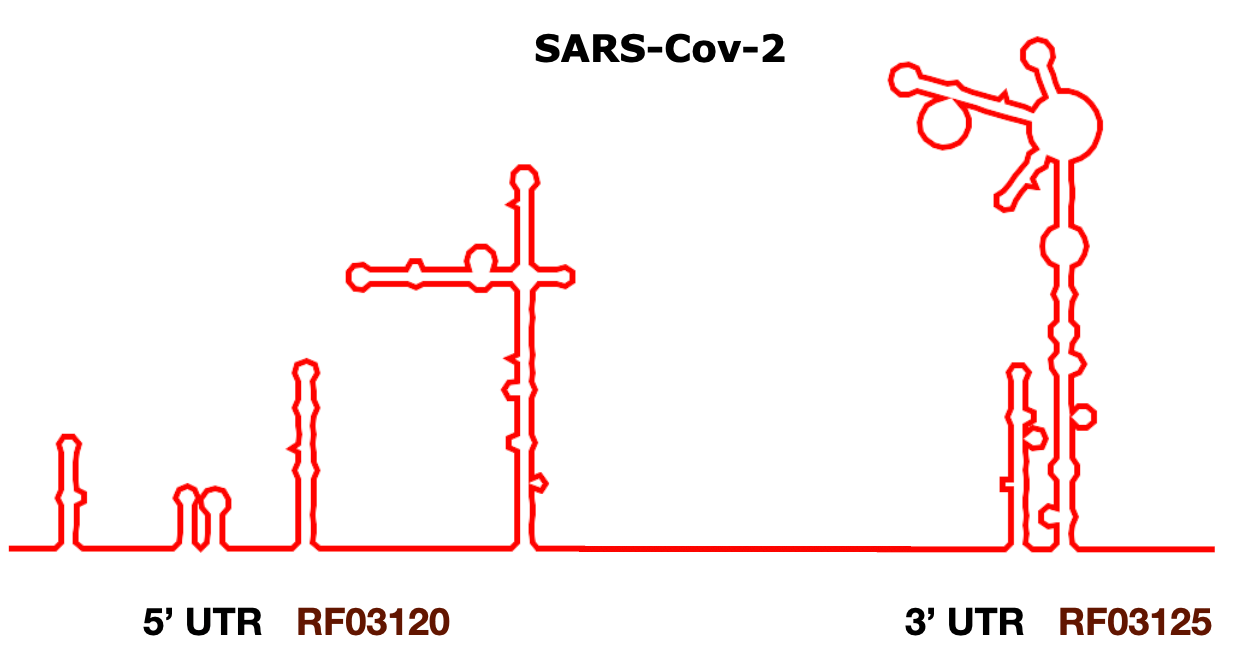
In Rfam release 14.2 we created 10 families representing the entire 5'- and 3'- untranslated regions (UTRs) for Alpha-, Beta-, Gamma-, and Deltacoronaviruses. A specialised set of alignments for the subgenus Sarbecovirus is also provided, including the SARS-CoV-1 and SARS-CoV-2 UTRs.
There are also 4 non-UTR families, including Coronavirus packaging signal, Coronavirus frameshifting stimulation element, Coronavirus s2m RNA, and Coronavirus 3'-UTR pseudoknot.
| Accession | Rfam ID | Description |
|---|---|---|
| RF00507 | Corona_FSE | Coronavirus frameshifting stimulation element |
| RF00164 | s2m | Coronavirus 3' stem-loop II-like motif (s2m) |
| RF00165 | Corona_pk3 | Coronavirus 3' UTR pseudoknot |
| RF03116 | aCoV-5UTR | Alphacoronavirus 5'UTR |
| RF03117 | bCoV-5UTR | Betacoronavirus 5'UTR |
| RF03118 | gCoV-5UTR | Gammacoronavirus 5'UTR |
| RF03119 | dCoV-5UTR | Deltacoronavirus 5'UTR |
| RF03120 | Sarbecovirus-5UTR | Sarbecovirus 5'UTR |
| RF03121 | aCoV-3UTR | Alphacoronavirus 3'UTR |
| RF03122 | bCoV-3UTR | Betacoronavirus 3'UTR |
| RF03123 | gCoV-3UTR | Gammacoronavirus 3'UTR |
| RF03124 | dCoV-3UTR | Deltacoronavirus 3'UTR |
| RF03125 | Sarbecovirus-3UTR | Sarbecovirus 3'UTR |
Hepacivirus RNA families
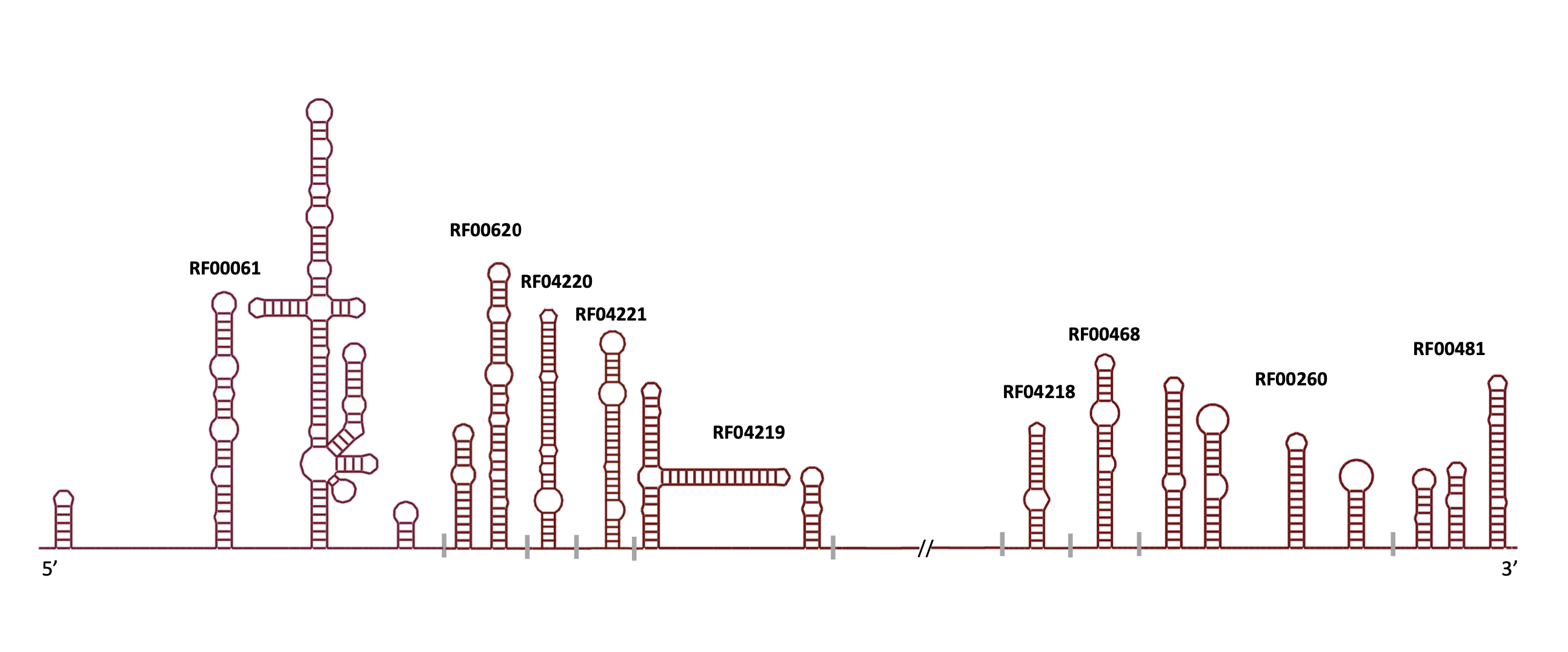
In release 14.10, we have updated 9 HCV families to better reflect their secondary structures.
In release 14.8, we have added 4 new Hepatitis C RNA families, and have updated 5 existing families. We have also removed 4 families: RF02585, RF02586, RF02587, RF02588.
| Accession | Rfam ID | Description |
|---|---|---|
| RF00061 | IRES_HCV | Hepatitis C virus internal ribosome entry site |
| RF00260 | HepC_CRE | Hepatitis C virus (HCV) cis-acting replication element (CRE) |
| RF00620 | HCV_ARF_SL | Hepatitis C alternative reading frame stem-loop |
| RF00468 | HCV_SLVII | Hepatitis C virus stem-loop VII |
| RF00481 | HCV_X3 | Hepatitis C virus 3'X element |
| RF04218 | HCV_5BSL1 | Hepatitis C virus stem-loop I |
| RF04219 | HCV_J750 | J750 non-coding RNA (containing SLSL761 and SL783) |
| RF04220 | HCV_SL588 | SL588 non-coding RNA |
| RF04221 | HCV_SL669 | SL669 non-coding RNA |
Flaviviridae RNA families
| Accession | Rfam ID | Description |
|---|---|---|
| RF03546 | Flavivirus-5UTR | Flavivirus 5' UTR |
| RF00185 | Flavi_CRE | Flavivirus 3' UTR cis-acting replication element (CRE) |
| RF00525 | Flavivirus_DB | Flavivirus DB element |
| RF03547 | Flavi_xrRNA | General Flavivirus exoribonuclease-resistant RNA element |
| RF03545 | Flavi_ISFV_CRE | Insect-specific Flavivirus 3' UTR cis-acting replication element (CRE) |
| RF03544 | Flavi_ISFV_repeat_Ra | Insect-Specific Flavivirus 3' UTR repeats Ra |
| RF03543 | Flavi_ISFV_repeat_Rb | Insect-Specific Flavivirus 3' UTR repeats Rb |
| RF03542 | Flavi_ISFV_repeat_Ra_Rb | Insect-Specific Flavivirus 3' UTR repeats Ra and Rb elements |
| RF03541 | Flavi_ISFV_xrRNA | Insect-specific Flavivirus exoribonuclease-resistant RNA element |
| RF03540 | Flavi_NKV_CRE | No-Known-Vector Flavivirus 3' UTR cis-acting replication element (CRE) |
| RF03539 | Flavi_NKV_xrRNA | No-known vector Flavivirus exoribonuclease-resistant RNA element |
| RF03538 | Flavi_TBFV_CRE | Tick-borne Flavivirus 3' UTR cis-acting replication element (CRE) |
| RF03537 | Flavi_TBFV_SL6 | Tick-borne Flavivirus short stem-loop SL6 |
| RF03536 | Flavi_TBFV_xrRNA | Tick-borne Flavivirus exoribonuclease-resistant RNA element | RF00209 | IRES_Pesti | Pestivirus internal ribosome entry site (IRES) including 5'UTR SLI |
| RF04322 | Pestivirus-3UTR-SLI | Pestivirus 3' UTR SLI |
| RF04323 | Pestivirus-3UTR-SLIV-SLII-BVDV-1 | Pestivirus 3' UTR SLIV, SLIII and SLII classical swine fever virus |
| RF04324 | Pestivirus-3UTR-SLIV-SLIII-SLII-CSFV | Pestivirus 3' UTR SLIV, SLIII and SLII classical swine fever virus |
| RF04325 | Pestivirus-3UTR-SLIII-scrofae | Pestivirus 3' UTR SLIII P. scrofae Atypical porcine pestivirus |
Work in progress
We are working on adding RNA families from other viruses, such as Filoviridae (e.g. Ebolavirus) and Rhabdoviridae (e.g. Rabies viruses).
Please contact Rfam if you are interested in a specific virus family or if you have feedback about this project.



