Summary
Wikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry Flavivirus 3' UTR. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
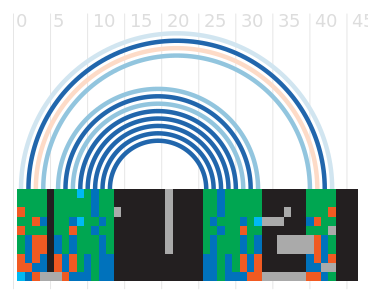
Alignment
Seed alignment view
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
Secondary structure
Species distribution
Sunburst controls
HideWeight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Gritsun TS, Gould EA Adv Virus Res. 2007;69:203-248. Origin and evolution of 3'UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. PUBMED:17222695
-
Gritsun DJ, Jones IM, Gould EA, Gritsun TS PLoS One. 2014;9:e92056. Molecular archaeology of Flaviviridae untranslated regions: duplicated RNA structures in the replication enhancer of flaviviruses and pestiviruses emerged via convergent evolution. PUBMED:24647143
-
Ochsenreiter R, Hofacker IL, Wolfinger MT Viruses. 2019; [Epub ahead of print] Functional RNA Structures in the 3'UTR of Tick-Borne, Insect-Specific and No-Known-Vector Flaviviruses. PUBMED:30909641
External database links
| Sequence Ontology: | SO:0000837 (UTR_region); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Lamkiewicz K | ||||||
| Structure source | Predicted; LocARNA 2.0.0RC8 | ||||||
| Type | Cis-reg; | ||||||
| Author |
Lamkiewicz K ,
Wolfinger MT ,
Wolfinger MT
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 30.00 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 38.3 |
| Trusted cutoff | 38.5 |
| Noise cutoff | 38.1 |
| Covariance model | Download |
 Loading...
Loading...