Summary
Wikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry Mir-275 microRNA precursor family. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
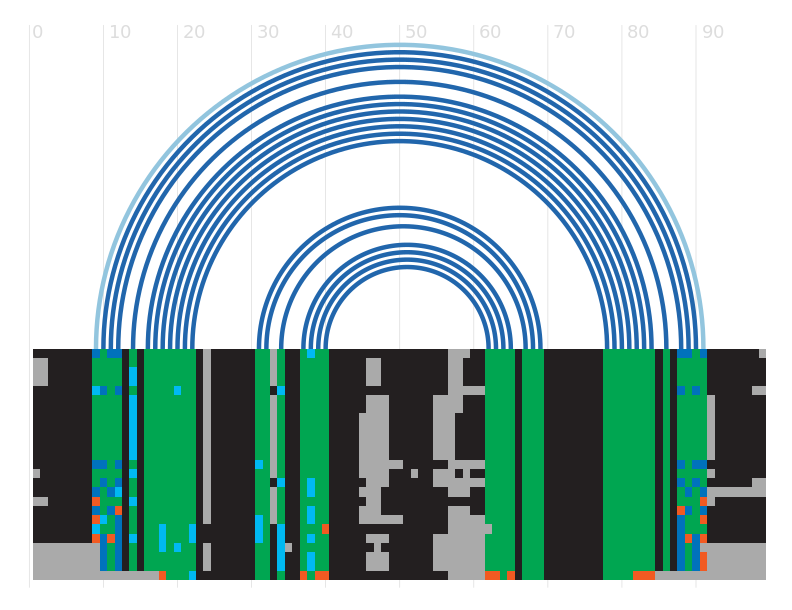
Alignment
Seed alignment view
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
Secondary structure
Species distribution
Sunburst controls
HideWeight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Winter F, Edaye S, Huttenhofer A, Brunel C Nucleic Acids Res. 2007;35:6953-6962. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. PUBMED:17933784
-
Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR BMC Genomics. 2010;11:119. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. PUBMED:20167119
-
Lai EC, Tomancak P, Williams RW, Rubin GM Genome Biol 2003;4:R42. Computational identification of Drosophila microRNA genes. PUBMED:12844358
-
Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T Dev Cell 2003;5:337-350. The small RNA profile during Drosophila melanogaster development. PUBMED:12919683
-
Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC Genome Res. 2007;17:1850-1864. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs PUBMED:17989254
-
Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M Genome Res. 2007;17:1865-1879. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes PUBMED:17989255
-
Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJ, Kitts P, Lynch JA, Murphy T, Oliveira DC, Smith CD, van de Zande L, Worley KC, Zdobnov EM, Aerts M, Albert S, Anaya VH, Anzola JM, Barchuk AR, Behura SK, Bera AN, Berenbaum MR, Bertossa RC, Bitondi MM, Bordenstein SR, Bork P, Bornberg-Bauer E, Brunain M, Cazzamali G, Chaboub L, Chacko J, Chavez D, Childers CP, Choi JH, Clark ME, Claudianos C, Clinton RA, Cree AG, Cristino AS, Dang PM, Darby AC, de Graaf DC, Devreese B, Dinh HH, Edwards R, Elango N, Elhaik E, Ermolaeva O, Evans JD, Foret S, Fowler GR, Gerlach D, Gibson JD, Gilbert DG, Graur D, Grunder S, Hagen DE, Han Y, Hauser F, Hultmark D, Hunter HC 4th, Hurst GD, Jhangian SN, Jiang H, Johnson RM, Jones AK, Junier T, Kadowaki T, Kamping A, Kapustin Y, Kechavarzi B, Kim J, Kim J, Kiryutin B, Koevoets T, Kovar CL, Kriventseva EV, Kucharski R, Lee H, Lee SL, Lees K, Lewis LR, Loehlin DW, Logsdon JM Jr, Lopez JA, Lozado RJ, Maglott D, Maleszka R, Mayampurath A, Mazur DJ, McClure MA, Moore AD, Morgan MB, Muller J, Munoz-Torres MC, Muzny DM, Nazareth LV, Neupert S, Nguyen NB, Nunes FM, Oakeshott JG, Okwuonu GO, Pannebakker BA, Pejaver VR, Peng Z, Pratt SC, Predel R, Pu LL, Ranson H, Raychoudhury R, Rechtsteiner A, Reese JT, Reid JG, Riddle M, Robertson HM, Romero-Severson J, Rosenberg M, Sackton TB, Sattelle DB, Schluns H, Schmitt T, Schneider M, Schuler A, Schurko AM, Shuker DM, Simoes ZL, Sinha S, Smith Z, Solovyev V, Souvorov A, Springauf A, Stafflinger E, Stage DE, Stanke M, Tanaka Y, Telschow A, Trent C, Vattathil S, Verhulst EC, Viljakainen L, Wanner KW, Waterhouse RM, Whitfield JB, Wilkes TE, Williamson M, Willis JH, Wolschin F, Wyder S, Yamada T, Yi SV, Zecher CN, Zhang L, Gibbs RA Science. 2010;327:343-348. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. PUBMED:20075255
-
Yu X, Zhou Q, Li SC, Luo Q, Cai Y, Lin WC, Chen H, Yang Y, Hu S, Yu J PLoS One. 2008;3:e2997. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PUBMED:18714353
-
Cao J, Tong C, Wu X, Lv J, Yang Z, Jin Y Insect Biochem Mol Biol. 2008;38:1066-1071. Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. PUBMED:18977439
-
Liu S, Li D, Li Q, Zhao P, Xiang Z, Xia Q BMC Genomics. 2010;11:148. MicroRNAs of Bombyx mori identified by Solexa sequencing. PUBMED:20199675
-
Yu X, Zhou Q, Cai Y, Luo Q, Lin H, Hu S, Yu J Genomics. 2009;94:438-444. A discovery of novel microRNAs in the silkworm (Bombyx mori) genome. PUBMED:19699294
-
Cai Y, Yu X, Zhou Q, Yu C, Hu H, Liu J, Lin H, Yang J, Zhang B, Cui P, Hu S, Yu J Funct Integr Genomics. 2010;10:405-415. Novel microRNAs in silkworm (Bombyx mori). PUBMED:20229201
-
Li S, Mead EA, Liang S, Tu Z BMC Genomics. 2009;10:581. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. PUBMED:19961592
-
Marco A, Hui JH, Ronshaugen M, Griffiths-Jones S Genome Biol Evol. 2010;2:686-696. Functional shifts in insect microRNA evolution. PUBMED:20817720
-
Kozomara A, Birgaoanu M, Griffiths-Jones S Nucleic Acids Res. 2019;47:D155. miRBase: from microRNA sequences to function. PUBMED:30423142
External database links
| Gene Ontology: | GO:0016442 (RISC complex); GO:0035195 (miRNA-mediated post-transcriptional gene silencing); |
| Sequence Ontology: | SO:0001244 (pre_miRNA); |
| MIPF: | MIPF0000187 |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Griffiths-Jones SR | ||||||
| Structure source | Predicted; RNAalifold | ||||||
| Type | Gene; miRNA; | ||||||
| Author |
Griffiths-Jones SR
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 30.00 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 56.0 |
| Trusted cutoff | 56.4 |
| Noise cutoff | 55.5 |
| Covariance model | Download |
 Loading...
Loading...