Summary
Note on Riboswitches
This Rfam family PreQ1 (RF00522) represents an aptamer domain of a full riboswitch PreQ1 (pre queuosine) riboswitch aptamer. Riboswitches are non-coding RNA structures that regulate gene expression in response to ligand. Each riboswitch has two main parts: the aptamer domain and the expression platform. The aptamer domain is highly conserved to precisely bind its ligand. However, the expression platform has multiple modes of gene regulation, which introduces sequence and structure variability that increases difficulty in its detection through covariance model searching. For more information see the original publications.
Wikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry PreQ1 riboswitch. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
Alignment
Seed alignment view
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
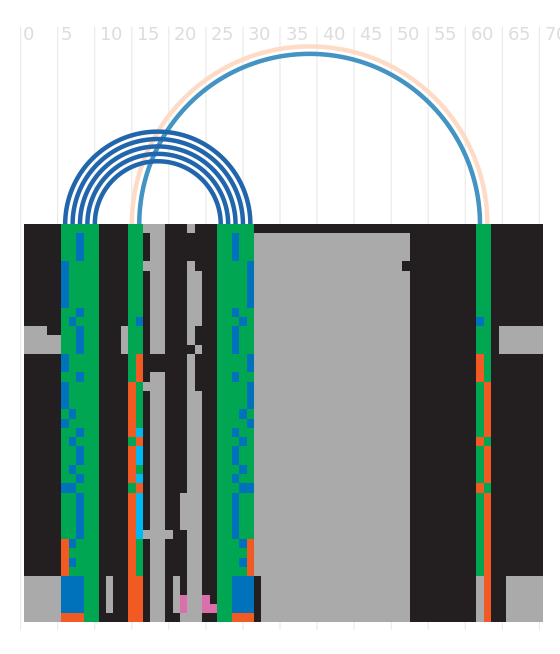
Secondary structure
Species distribution
Sunburst controls
HideWeight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Reader JS, Metzgar D, Schimmel P, de Crecy-Lagard V J Biol Chem 2004;279:6280-6285. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. PUBMED:14660578
-
Kang M, Peterson R, Feigon J Mol Cell. 2009;33:784-790. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. PUBMED:19285444
-
Flemmich L, Heel S, Moreno S, Breuker K, Micura R Nat Commun. 2021;12:3877. A natural riboswitch scaffold with self-methylation activity. PUBMED:34162884
-
Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE J Biol Chem. 2009;284:11012-11016. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. PUBMED:19261617
-
Klein DJ, Edwards TE, Ferre-D'Amare AR Nat Struct Mol Biol. 2009;16:343-344. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. PUBMED:19234468
-
Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE J Biol Chem. 2011;286:24626-24637. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. PUBMED:21592962
-
Connelly CM, Numata T, Boer RE, Moon MH, Sinniah RS, Barchi JJ, Ferre-D'Amare AR, Schneekloth JS Jr Nat Commun. 2019;10:1501. Synthetic ligands for PreQ(1) riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure. PUBMED:30940810
-
Schroeder GM, Dutta D, Cavender CE, Jenkins JL, Pritchett EM, Baker CD, Ashton JM, Mathews DH, Wedekind JE Nucleic Acids Res. 2020;48:8146-8164. Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation. PUBMED:32597951
-
Balaratnam S, Rhodes C, Bume DD, Connelly C, Lai CC, Kelley JA, Yazdani K, Homan PJ, Incarnato D, Numata T, Schneekloth JS Jr Nat Commun. 2021;12:5856. A chemical probe based on the PreQ(1) metabolite enables transcriptome-wide mapping of binding sites. PUBMED:34615874
-
Schroeder GM, Cavender CE, Blau ME, Jenkins JL, Mathews DH, Wedekind JE Nat Commun. 2022;13:199. A small RNA that cooperatively senses two stacked metabolites in one pocket for gene control. PUBMED:35017488
-
Schroeder GM, Akinyemi O, Malik J, Focht CM, Pritchett EM, Baker CD, McSally JP, Jenkins JL, Mathews DH, Wedekind JE Nucleic Acids Res. 2023;51:2464-2484. A riboswitch separated from its ribosome-binding site still regulates translation. PUBMED:36762498
-
Flemmich L, Heel S, Moreno S, Breuker K, Micura R Nat Commun. 2021;12:3877. A natural riboswitch scaffold with self-methylation activity. PUBMED:34162884
External database links
| Gene Ontology: | GO:0008616 (queuosine biosynthetic process); |
| Sequence Ontology: | SO:0000035 (riboswitch); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Barrick JE | ||||||
| Structure source | Predicted; Barrick JE | ||||||
| Type | Cis-reg; riboswitch; | ||||||
| Author |
Barrick JE,
Breaker RR ,
Ontiveros-Palacios N ,
Ontiveros-Palacios N
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 30.00 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 39.0 |
| Trusted cutoff | 39.0 |
| Noise cutoff | 38.9 |
| Covariance model | Download |
 Loading...
Loading...