Summary
Wikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry Cripavirus internal ribosome entry site (IRES). More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
Alignment
Seed alignment view
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
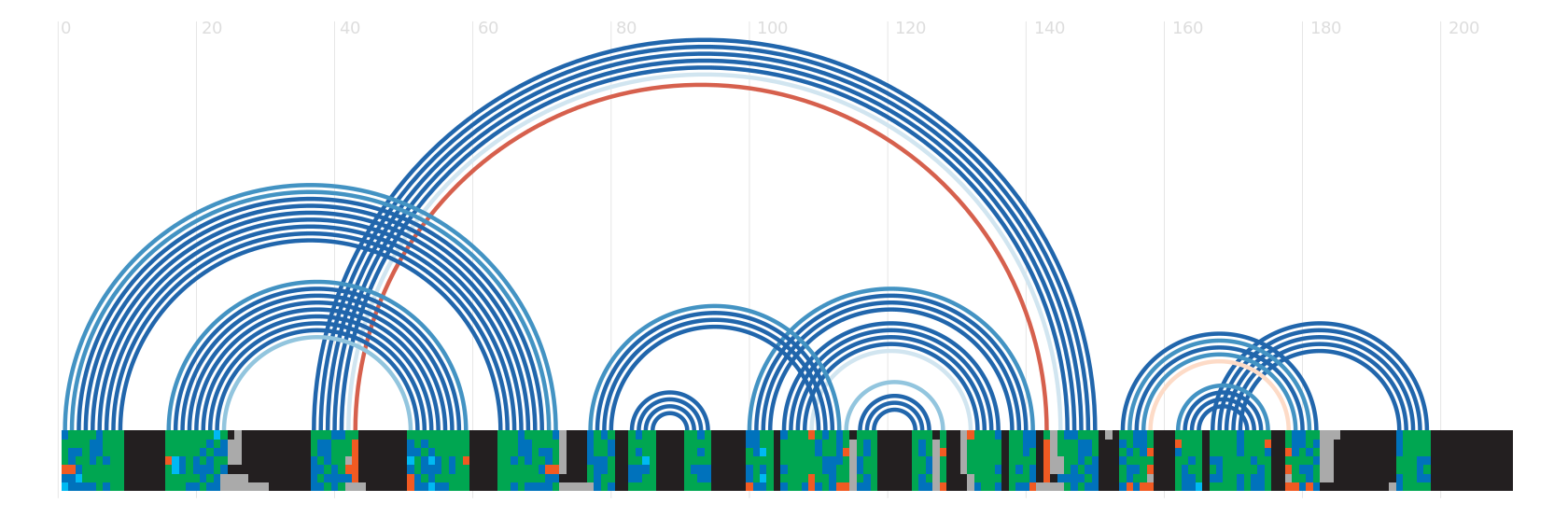
Secondary structure
Species distribution
Sunburst controls
HideWeight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
Structures
For those sequences which have a structure in the Protein DataBank, we generate a mapping between EMBL, PDB and Rfam coordinate systems. The table below shows the structures on which the IRES_Cripavirus family has been found.
Loading structure mapping...
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Kanamori Y, Nakashima N RNA 2001;7:266-274. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. PUBMED:11233983
-
Pfingsten JS, Costantino DA, Kieft JS Science. 2006;314:1450-1454. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. PUBMED:17124290
-
Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V Cell. 2014;157:823-831. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. PUBMED:24792965
-
Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM Mol Cell. 2015;57:422-432. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. PUBMED:25601755
-
Colussi TM, Costantino DA, Zhu J, Donohue JP, Korostelev AA, Jaafar ZA, Plank TD, Noller HF, Kieft JS Nature. 2015;519:110-113. Initiation of translation in bacteria by a structured eukaryotic IRES RNA. PUBMED:25652826
-
Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernandez IS Elife. 2016;5:e13567. Structural characterization of ribosome recruitment and translocation by type IV IRES. PUBMED:27159451
-
Pisareva VP, Pisarev AV, Fernandez IS Elife. 2018;7:e34062. Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES. PUBMED:29856316
-
Yuan S, Peng L, Park JJ, Hu Y, Devarkar SC, Dong MB, Shen Q, Wu S, Chen S, Lomakin IB, Xiong Y Mol Cell. 2020;80:1055-1066. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. PUBMED:33188728
-
Hilal T, Killam BY, Grozdanovic M, Dobosz-Bartoszek M, Loerke J, Burger J, Mielke T, Copeland PR, Simonovic M, Spahn CMT Science. 2022;376:1338-1343. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. PUBMED:35709277
-
Kogel A, Keidel A, Loukeri MJ, Kuhn CC, Langer LM, Schafer IB, Conti E Nature. 2024;635:237-242. Structural basis of mRNA decay by the human exosome-ribosome supercomplex. PUBMED:39385025
External database links
| Gene Ontology: | GO:0043022 (ribosome binding); |
| Sequence Ontology: | SO:0000243 (internal_ribosome_entry_site); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Published; 11233983 | ||||||
| Structure source | Published; PMID:11233983 | ||||||
| Type | Cis-reg; IRES; | ||||||
| Author |
Moxon SJ ,
Ontiveros-Palacios N ,
Ontiveros-Palacios N
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 30.00 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 44.5 |
| Trusted cutoff | 44.5 |
| Noise cutoff | 41.2 |
| Covariance model | Download |
 Loading...
Loading...